Our Vision
Tailoring therapies to an individual’s unique molecular profile is essential for effective treatment, and this requires a deep understanding of the cellular and macromolecular mechanisms that influence treatment response. Biologists have long sought to simulate the state and functioning of cells in order to understand and control their core processes. Recent advances in high-throughput technologies that screen biological systems at an unprecedented resolution have opened new avenues for machine learning approaches to tackle these challenges successfully.
Our lab develops novel artificial intelligence methods designed to mimic the structure and inner workings of biological systems, allowing them to be trained on large biomedical datasets from various experimental technologies. By leveraging these AI techniques inspired by biological systems, our work not only provides important contributions to personalized medicine but also impacts active machine learning research. As we unlock a deeper understanding of cellular mechanisms, we aim to enable more effective and individualized therapies for patients.
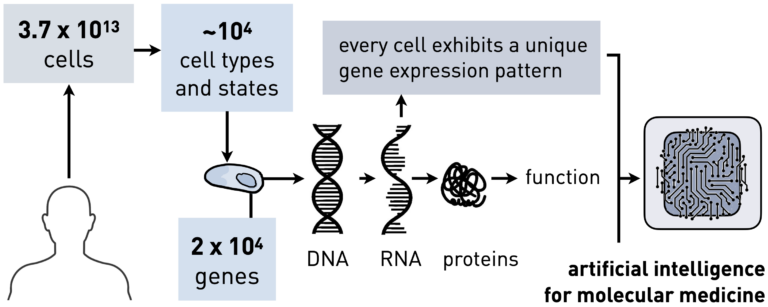
Neural Network Architectures for Biomedicine
To advance personalized medicine, we need to design deep learning architectures that incorporate biomedical priors, scale to large biological datasets, and are deployable in the clinical setting. Such architectures will be essential for making reliable predictions of therapy outcomes[1] and forecasting the molecular behavior of different drugs[2][3].
Recent Highlights
On Building and Designing Foundation Models
From association to reasoning: foundation models must learn to ask “what if.” An essay on their design and building blocks.
How to Build the AI Virtual Cell
Our vision on how to build multi-scale multi-modal foundation models for biomedicine.
Tutorial on Neural OT and Diffusion Models
A tutorial on neural optimal transport to diffusion models: theory and applications.
Our previous work has developed AI methods, such as diffusion models[4] and neural optimal transport[5][6], to learn and infer dynamic responses of single cells derived from cancer patients using both imaging and sequencing data. By integrating insights of the underlying biological mechanisms with concepts from dynamic systems theory, and contributing novel theoretical and algorithmic frameworks, these tools can forecast the dynamics of complex biological processes. For a thorough review of these methods and the related theory, refer to our tutorial on neural optimal transport and diffusion models presented at the International Conference on Machine Learning (ICML).
Similarly, in order to incorporate physical and geometrical first principles for learning with macromolecular structures and their interactions, our past research developed graph[7] and equivariant neural networks[8] that are able to infer binding mechanisms and docking between molecules and proteins. This is crucial in particular for drug design, e.g., to obtain a better understanding of drug’s interactions with a target of interest.
But this has only been the starting point: The increasing resolution of experimental technologies as well as the growing complexity and size of biological datasets provides an even greater need and opportunity for active research on machine learning architectures that tackle the aforementioned challenges. So, stay tuned for future work and if interested, please reach out!
Personalized Treatments and Therapy Design
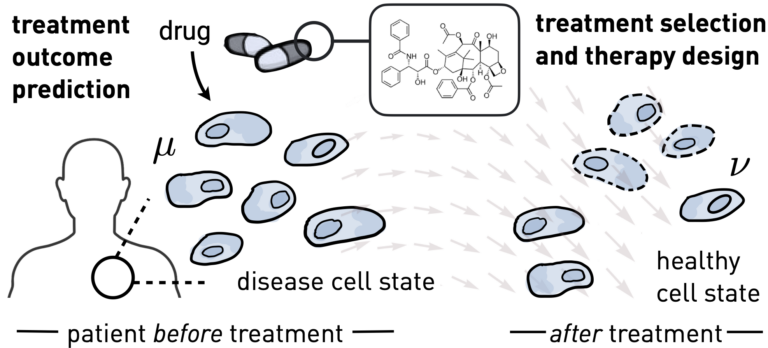
Predicting responses of heterogeneous cell populations to molecular perturbations (e.g., genetic knockouts, chemical drugs, or developmental signals) at the level of single cells is crucial for deciphering molecular processes and obtaining a better understanding of function and disease. Our efforts in that area[9] directly translate into clinical applications: In a current observational clinical study of cancer patients within the Tumor Profiler consortium, our tools demonstrate the potential for meaningful treatment outcome predictions for a diverse set of patients.
The evolving landscape of clinical data of different modalities opens up numerous avenues for future research in artificial intelligence. By combining data from genomics, proteomics, and other omics with clinical data, AI can build holistic models of disease and treatment response and identify new therapeutic targets based on patient-specific data and predicted drug responses. Our research in this field can thus significantly advance our understanding of diseases and improve patient outcomes through more precise and personalized medical interventions.
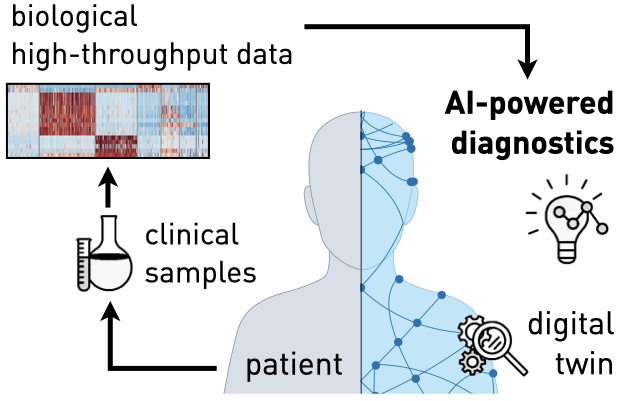
Digital Diagnostics and the Vision of the Digital Twin
Building on insights from AI breakthroughs in vision and language and leveraging vast amounts of biological data, our current research aims to advance the vision of a virtual cell, a foundational AI-based model capable of predicting the behavior of healthy and diseased cells with broad applications for biomedical research and therapeutic development. AI virtual cells allow the creation of a patient’s medical digital twin, leading to faster diagnoses, more successful treatments, and the detection of rare diseases. Such machine learning models then allow simulating patient-specific treatment responses and providing an in silico environment for testing drug candidates and conduct diagnosis of diseases.
Creating such virtual patient representations as well as digital diagnostics requires innovations on adaptive algorithms that navigate the constraints of clinical settings and the puzzling complexity of human biology.
References
- Charlotte Bunne and Stefan Stark and Gabriele Gut and … and Mitchell Levesque and Kjong Van Lehmann and Lucas Pelkmans and Andreas Krause and Gunnar Rätsch (2023): Learning Single-Cell Perturbation Responses using Neural Optimal Transport. In: Nature Methods, 2023, (Highlighted as Research Briefing in Nature Methods. Also presented at the NeurIPS Workshop on Optimal Transport and Machine Learning (OTML), 2021).
- Octavian-Eugen Ganea and Xinyuan Huang and Charlotte Bunne and Yatao Bian and Regina Barzilay and Tommi Jaakkola and Andreas Krause (2022): Independent SE (3)-Equivariant Models for End-to-End Rigid Protein Docking. In: International Conference on Learning Representations (ICLR), 2022, (Spotlight Talk at ICLR and Ranked Top 15 among 3326 Submissions (Top 0.4 percent) and Contributed Talk at ELLIS Machine Learning for Molecule Discovery Workshop, 2021).
- Charlotte Bunne and Vignesh Ram Somnath and Andreas Krause (2021): Multi-Scale Representation Learning on Proteins. In: Advances in Neural Information Processing Systems (NeurIPS), 2021, (Spotlight Talk at ICML Computational Biology Workshop, 2021).
- Vignesh Ram Somnath and Matteo Pariset and Ya-Ping Hsieh and Maria Rodriguez Martinez and Andreas Krause and Charlotte Bunne (2023): Aligned Diffusion Schrödinger Bridges. In: Conference on Uncertainty in Artificial Intelligence (UAI), 2023, (Also presented at the ICML Workshop on New Frontiers for Learning, Control, and Dynamical Systems, 2023).
- Charlotte Bunne and Andreas Krause and Marco Cuturi (2022): Supervised Training of Conditional Monge Maps. In: Advances in Neural Information Processing Systems (NeurIPS), 2022, (Also presented at the ICML Workshop on Interpretable Machine Learning in Healthcare (IMLH), 2022).
- Charlotte Bunne and Laetitia Meng-Papaxanthos and Andreas Krause and Marco Cuturi (2022): Proximal Optimal Transport Modeling of Population Dynamics. In: International Conference on Artificial Intelligence and Statistics (AISTATS), 2022, (Best Paper Award and Contributed Talk at ICML Time Series Workshop, 2021).
- Charlotte Bunne and Vignesh Ram Somnath and Andreas Krause (2021): Multi-Scale Representation Learning on Proteins. In: Advances in Neural Information Processing Systems (NeurIPS), 2021, (Spotlight Talk at ICML Computational Biology Workshop, 2021).
- Octavian-Eugen Ganea and Xinyuan Huang and Charlotte Bunne and Yatao Bian and Regina Barzilay and Tommi Jaakkola and Andreas Krause (2022): Independent SE (3)-Equivariant Models for End-to-End Rigid Protein Docking. In: International Conference on Learning Representations (ICLR), 2022, (Spotlight Talk at ICLR and Ranked Top 15 among 3326 Submissions (Top 0.4 percent) and Contributed Talk at ELLIS Machine Learning for Molecule Discovery Workshop, 2021).
- Charlotte Bunne and Stefan Stark and Gabriele Gut and … and Mitchell Levesque and Kjong Van Lehmann and Lucas Pelkmans and Andreas Krause and Gunnar Rätsch (2023): Learning Single-Cell Perturbation Responses using Neural Optimal Transport. In: Nature Methods, 2023, (Highlighted as Research Briefing in Nature Methods. Also presented at the NeurIPS Workshop on Optimal Transport and Machine Learning (OTML), 2021).
